文章目录
前言
Frangi滤波原文:https://www.researchgate.net/publication/2388170_Multiscale_Vessel_Enhancement_Filtering
Frangi滤波翻译讲解:
https://zhuanlan.zhihu.com/p/127951058
参考代码:https://github.com/vinhnguyen21/python_JermanEnhancementFilter
Frangi滤波原文中详细说明了3D,2D图像下的血管强化方法,但是在网上找了好久,只有找到2D滤波的代码,在做毕设的时候因为时间有限,所以对三正交平面都进行一次2D Frangi滤波的方式代替3D Frangi滤波,虽然也有效果但总是不是很舒服。
本文首先会根据参考代码中的2D Frangi滤波进行讲解,接着在此基础上按照原文的意思更改3D Frangi滤波,最后放上几张结果图进行对比。
本人水平有限,还望各位大佬批评指正。
一、2D Frangi滤波——原文复现
1、import
需要说明的是,我们的3D文件是.nii文件,这里使用SimpleITK进行读写
import cv2
import os
import numpy as np
from scipy import ndimage
import SimpleITK as sitk
2、vesselness2d
vesselness2d.py(记得import上面的内容)
class vesselness2d:
def __init__(self, image, sigma, spacing, tau):super(vesselness2d, self).__init__()#image为numpy类型,表示n * m 的二维矩阵。
self.image = image
#sigma为list 类型,表示高斯核的尺度。
self.sigma = sigma
#spacing为list类型,表示.nii文件下某一切面下的体素的二维尺寸。如果输入图像本身为二维图像,则为[1,1],如果为三维图像,则为对应的space。
self.spacing = spacing
#tau为float类型,表示比例系数。
self.tau = tau
# 图像尺寸
self.size = image.shape
# 使用特定的特定sigma尺寸下的高斯核对图像滤波
# 这里作者并没有使用n*n的卷积核,而是分别使用n*1,1*n的卷积对图像进行x和y方向上的卷积,
# 并且使用的是最原始的计算高斯函数得到卷积核,而不是直接用现成的高斯卷积核,
# 通过证明可以发现在两方面的结果是等价的。
def gaussian(self, image, sigma):
siz = sigma *6 # 核的尺寸
#x轴方向上的滤波
temp =round(siz / self.spacing[0]/2)
x =[i for i in range(-temp, temp +1)]
x = np.array(x)
H = np.exp(-(x **2/(2*((sigma / self.spacing[0])**2))))
H = H / np.sum(H)
Hx = H.reshape(len(H),1,1)
I = ndimage.filters.convolve(image, Hx, mode='nearest')#y轴方向上的滤波
temp =round(siz / self.spacing[1]/2)
x =[i for i in range(-temp, temp +1)]
x = np.array(x)
H = np.exp(-(x **2/(2*((sigma / self.spacing[1])**2))))
H = H / np.sum(H[:])
Hy = H.reshape(1,len(H),1)
I = ndimage.filters.convolve(I, Hy, mode='nearest')return I
# 求图像的梯度
def gradient2(self, F, option):
k = self.size[0]
l = self.size[1]
D = np.zeros(F.shape)if option =="x":
D[0,:]= F[1,:]- F[0,:]
D[k -1,:]= F[k -1,:]- F[k -2,:]#takecenter differences on interior points
D[1:k -2,:]=(F[2:k -1,:]- F[0:k -3,:])/2else:
D[:,0]= F[:,1]- F[:,0]
D[:, l -1]= F[:, l -1]- F[:, l -2]
D[:,1:l -2]=(F[:,2:l -1]- F[:,0:l -3])/2return D
# 求海森矩阵中所需要的二阶偏导数
def Hessian2d(self, image, sigma):
image = self.gaussian(image, sigma)#image= ndimage.gaussian_filter(image, sigma, mode ='nearest')
Dy = self.gradient2(image,"y")
Dyy = self.gradient2(Dy,"y")
Dx = self.gradient2(image,"x")
Dxx = self.gradient2(Dx,"x")
Dxy = self.gradient2(Dx,'y')return Dxx, Dyy, Dxy
# 求解海森矩阵的两个特征值
# 这里作者使用求根公式,将二阶海森矩阵展开,a=1,b=-(Ixx+Iyy),c=(Ixx*Iyy-Ixy*Ixy)
# 首先计算 sqrt(b^2-4ac),通过化简得到tmp
# 最后得到两个特征值mu1,mu2,根据大小关系,大的为mu2,小的为mu1
def eigvalOfhessian2d(self, Dxx, Dyy, Dxy):
tmp = np.sqrt((Dxx - Dyy)**2+4*(Dxy **2))#computeeigenvectors of J, v1 and v2
mu1 =0.5*(Dxx + Dyy + tmp)
mu2 =0.5*(Dxx + Dyy - tmp)#Sort eigen values by absolute value abs(Lambda1)<abs(Lambda2)
indices =(np.absolute(mu1)> np.absolute(mu2))
Lambda1 = mu1
Lambda1[indices]= mu2[indices]
Lambda2 = mu2
Lambda2[indices]= mu1[indices]return Lambda1, Lambda2
def imageEigenvalues(self, I, sigma):
hxx, hyy, hxy = self.Hessian2d(I, sigma)
c = sigma **2
hxx =-c * hxx
hyy =-c * hyy
hxy =-c * hxy
# 为了降低运算量,去掉噪声项的计算
B1 =-(hxx + hyy)
B2 = hxx * hyy - hxy **2
T = np.ones(B1.shape)
T[(B1 <0)]=0
T[(B1 ==0)&(B2 ==0)]=0
T = T.flatten()
indeces = np.where(T ==1)[0]
hxx = hxx.flatten()
hyy = hyy.flatten()
hxy = hxy.flatten()
hxx = hxx[indeces]
hyy = hyy[indeces]
hxy = hxy[indeces]
lambda1i, lambda2i = self.eigvalOfhessian2d(hxx, hyy, hxy)
lambda1 = np.zeros(self.size[0]* self.size[1],)
lambda2 = np.zeros(self.size[0]* self.size[1],)
lambda1[indeces]= lambda1i
lambda2[indeces]= lambda2i
# 去掉噪声
lambda1[(np.isinf(lambda1))]=0
lambda2[(np.isinf(lambda2))]=0
lambda1[(np.absolute(lambda1)<1e-4)]=0
lambda1 = lambda1.reshape(self.size)
lambda2[(np.absolute(lambda2)<1e-4)]=0
lambda2 = lambda2.reshape(self.size)return lambda1, lambda2
# 血管强化
def vesselness2d(self):for j in range(len(self.sigma)):
lambda1, lambda2 = self.imageEigenvalues(self.image, self.sigma[j])
lambda3 = lambda2.copy()
new_tau = self.tau * np.min(lambda3)
lambda3[(lambda3 <0)&(lambda3 >= new_tau)]= new_tau
different = lambda3 - lambda2
response =((np.absolute(lambda2)**2)* np.absolute(different))*27/((2* np.absolute(lambda2)+ np.absolute(different))**3)
response[(lambda2 < lambda3 /2)]=1
response[(lambda2 >=0)]=0
response[np.where(np.isinf(response))[0]]=0if j ==0:
vesselness = response
else:
vesselness = np.maximum(vesselness, response)
vesselness[(vesselness <1e-2)]=0return vesselness
3、应用示例(原文)
demo.py
需要说明的是,这里使用的图像是0-255灰度图像,原文的强化针对的是背景亮,血管暗的图像,但是这里的图像是相反,所以在下面对图像进行了像素灰度值的反转。
from PIL import Image
import numpy as np
import cv2
import matplotlib.pyplot as plt
from vesselness2d import *
img_dir ='images/test.tif' #路径写自己的
#readingimage
image = Image.open(img_dir).convert("RGB")
image = np.array(image)
plt.figure(figsize=(10,10))
plt.imshow(image, cmap='gray')#convertforgeground to background and vice-versa
image =255-image
image = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)
thr = np.percentile(image[(image >0)],1)*0.9
image[(image <= thr)]= thr
image = image - np.min(image)
image = image / np.max(image)
sigma=[0.5,1,1.5,2,2.5]
spacing =[1,1]
tau =2
output =vesselness2d(image, sigma, spacing, tau)
output = output.vesselness2d()
plt.figure(figsize=(10,10))
plt.imshow(output, cmap='gray')
原图:

结果: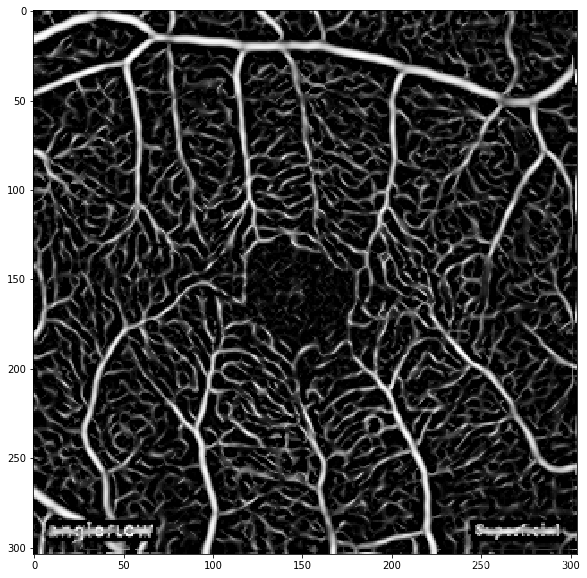
二、3D Frangi滤波 ——三正交平面分别进行2D Frangi滤波
1、import
import cv2 as cv
import SimpleITK as sitk
from vesselness2d import *
2、main
Hessian_3D.py
# 这里使用的是MSD数据集中的肝脏血管分割数据集,并且只用已训练好的肝脏分割模型对其进行分割,
# 只保留肝脏区域,图像的灰度范围是[0,200],血管相较于背景为白色
def edge(img,position):
img_dt = np.zeros((len(img),len(img[0]),len(img[0][0])))
img_dt[:]= img[:]
origin = img_dt[0][0][0]
img_dt[img_dt!=-origin]=1
img_dt[img_dt==-origin]=0
tmp = np.ones((len(img_dt),len(img_dt[0]),len(img_dt[0][0])))if position =="x":for i in range(len(img_dt)):
kernel = cv.getStructuringElement(cv.MORPH_RECT,(3,3))
dst = cv.erode(img_dt[i], kernel)
tmp[i]= dst
img_dt[tmp ==1]=0
elif position =="y":for i in range(len(img_dt[0])):
kernel = cv.getStructuringElement(cv.MORPH_RECT,(3,3))
dst = cv.erode(img_dt[:,i,:], kernel)
tmp[:,i,:]= dst
img_dt[tmp ==1]=0
elif position =="z":for i in range(len(img_dt[0][0])):
kernel = cv.getStructuringElement(cv.MORPH_RECT,(3,3))
dst = cv.erode(img_dt[:,:,i], kernel)
tmp[:,:,i]= dst
img_dt[tmp ==1]=0return img_dt
def frangi(img, sigma, spacing, tau,position):
img_dt = np.zeros((len(img),len(img[0]),len(img[0][0])))
img_dt[:]= img[:]
result_dt = np.zeros((len(img_dt),len(img_dt[0]),len(img_dt[0][0])))if position =="x":for i in range(len(img_dt)):
image = img_dt[i]
output =vesselness2d(image, sigma, spacing, tau)
output = output.vesselness2d()
result_dt[i]= output
elif position =="y":for i in range(len(img_dt[0])):
image = img_dt[:,i,:]
output =vesselness2d(image, sigma, spacing, tau)
output = output.vesselness2d()
result_dt[:,i,:]= output
elif position =="z":for i in range(len(img_dt[0][0])):
image = img_dt[:,:,i]
output =vesselness2d(image, sigma, spacing, tau)
output = output.vesselness2d()
result_dt[:,:,i]= output
return result_dt
def Hessian3D(image,sigma, tau):
img_dt = sitk.GetArrayFromImage(image)
stand = img_dt[0][0][0]
img_dt[img_dt==stand]=-200
img_dt =200-img_dt
img_dt[img_dt==400]=-200
edge_x =edge(img_dt,"x")
edge_y =edge(img_dt,"y")
edge_z =edge(img_dt,"z")
edge_x[edge_y ==1]=1
edge_x[edge_z ==1]=1
space = image.GetSpacing()
spacing_x =[space[0],space[1]]
spacing_y =[space[0],space[2]]
spacing_z =[space[1],space[2]]
hessian_x =frangi(img_dt, sigma, spacing_x, tau,"x")return
hessian_y =frangi(img_dt, sigma, spacing_y, tau,"y")
hessian_z =frangi(img_dt, sigma, spacing_z, tau,"z")
result_dt = hessian_x+hessian_y+hessian_z
result_dt[-1]= np.zeros((len(result_dt[0]),len(result_dt[0][0])))
result_dt[edge ==1]=0
result_dt *=400
result_dt[result_dt >200]=200
result_dt[img_dt ==-200]=-200
result_dt = result_dt.astype(int)
result = sitk.GetImageFromArray(result_dt)
result.SetSpacing(image.GetSpacing())
result.SetOrigin(image.GetOrigin())
result.SetDirection(image.GetDirection())return result
# 这里的main函数根据自己的需要改
# 这里我的直接对整个文件夹中的全部.nii文件进行处理
if __name__ =="__main__":
sigma =[0.5,1,1.5,2,2.5]
tau =2
path ="D:\\PythonProject\\Daily\\AHE"
result_path ="F:\\3DUNet-Pytorch-master_vesselSeg\\raw_dataset\\train_seg\\hessian"
path_list = os.listdir(path)for i in path_list:
image_i_path = os.path.join(path,i)
img = sitk.ReadImage(image_i_path)
result =Hessian3D(img,sigma,tau)
sitk.WriteImage(result,os.path.join(result_path,i))print(i +" is OK!")
原图:
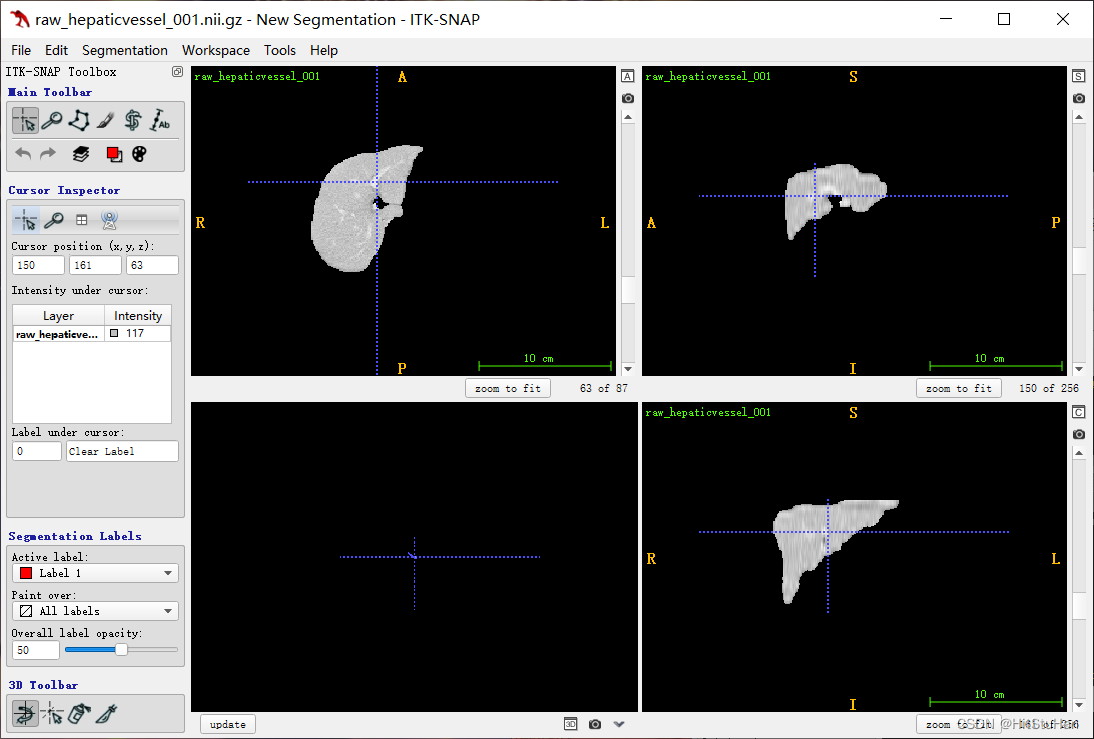
结果:
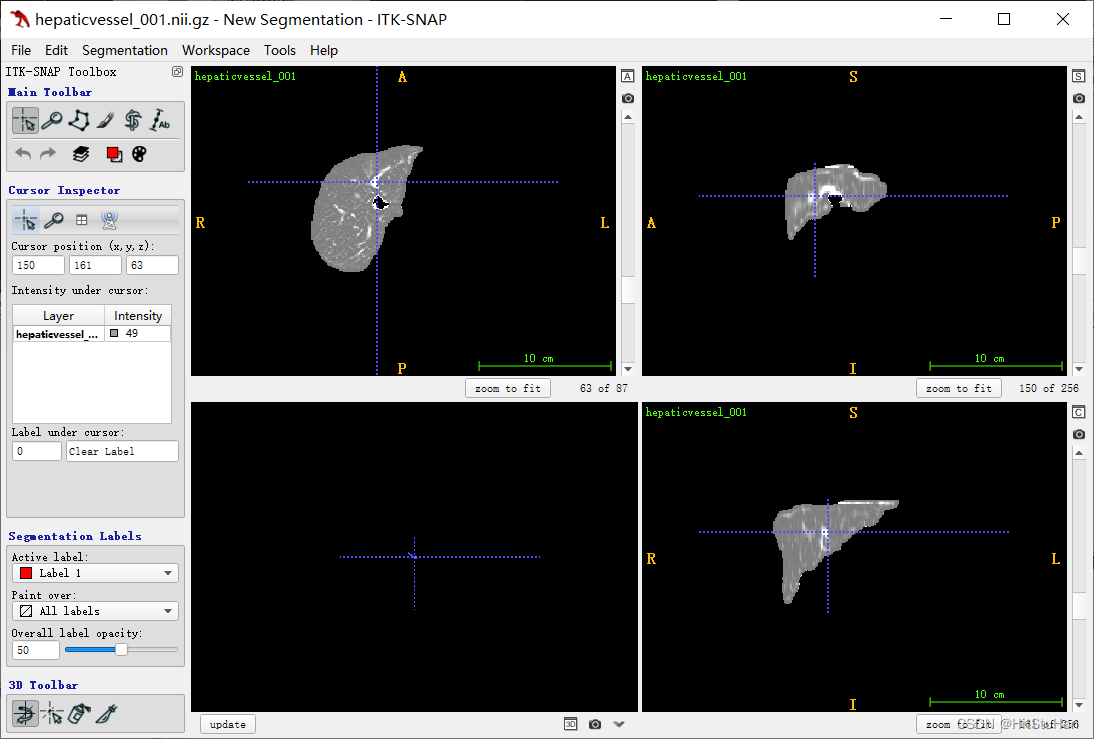
三、3D Frangi滤波 ——原文复现
1、import
import cv2 as cv
import SimpleITK as sitk
from vesselness2d import *
2、vesselness3d
Hessian_3D.py
# 对于3D Frangi滤波,与2D Frangi不同点在于
# 1、高斯滤波考虑第三个维度
# 2、构造三阶海森矩阵,[[Ixx,Ixy,Ixz],[Ixy,Iyy,Iyz],[Ixz,Iyz,Izz]]
# 3、求解三阶海森矩阵的特征值lambda1,lambda2,lambda3,并按照绝对值的大小排序
# 4、为减小求解时间,对于Ixx+Iyy+Izz<0的情况直接将灰度置为0
# 5、使用三维滤波公式求解体素灰度值
class vesselness3d:
def __init__(self, image, sigma, spacing):super(vesselness3d, self).__init__()
self.image = image
self.sigma = sigma
self.spacing = spacing
self.size = image.shape
def gaussian(self, image, sigma):
siz = sigma *6
temp =round(siz / self.spacing[0]/2)#processingx-axis
x =[i for i in range(-temp, temp +1)]
x = np.array(x)
H = np.exp(-(x **2/(2*((sigma / self.spacing[0])**2))))
H = H / np.sum(H)
Hx = H.reshape(len(H),1,1)
I = ndimage.filters.convolve(image, Hx, mode='nearest')#processingy-axis
temp =round(siz / self.spacing[1]/2)
x =[i for i in range(-temp, temp +1)]
x = np.array(x)
H = np.exp(-(x **2/(2*((sigma / self.spacing[1])**2))))
H = H / np.sum(H[:])
Hy = H.reshape(1,len(H),1)
I = ndimage.filters.convolve(I, Hy, mode='nearest')#processingz-axis
temp =round(siz / self.spacing[2]/2)
x =[i for i in range(-temp, temp +1)]
x = np.array(x)
H = np.exp(-(x **2/(2*((sigma / self.spacing[2])**2))))
H = H / np.sum(H[:])
Hz = H.reshape(1,1,len(H))
I = ndimage.filters.convolve(I, Hz, mode='nearest')return I
def gradient2(self, F, option):
k = self.size[0]
l = self.size[1]
h = self.size[2]
D = np.zeros(F.shape)if option =="x":
D[0,:,:]= F[1,:,:]- F[0,:,:]
D[k -1,:,:]= F[k -1,:,:]- F[k -2,:,:]#takecenter differences on interior points
D[1:k -2,:,:]=(F[2:k -1,:,:]- F[0:k -3,:,:])/2
elif option =="y":
D[:,0,:]= F[:,1,:]- F[:,0,:]
D[:, l -1,:]= F[:, l -1,:]- F[:, l -2,:]
D[:,1:l -2,:]=(F[:,2:l -1,:]- F[:,0:l -3,:])/2
elif option =="z":
D[:,:,0]= F[:,:,1]- F[:,:,0]
D[:,:, h-1]= F[:,:, h -1]- F[:,:, h -2]
D[:,:,1:h -2]=(F[:,:,2:h -1]- F[:,:,0:h -3])/2return D
def Hessian2d(self, image, sigma):
image = self.gaussian(image, sigma)
self.gaus_image = image
Dz = self.gradient2(image,"z")
Dzz = self.gradient2(Dz,"z")
Dy = self.gradient2(image,"y")
Dyy = self.gradient2(Dy,"y")
Dyz = self.gradient2(Dy,"z")
Dx = self.gradient2(image,"x")
Dxx = self.gradient2(Dx,"x")
Dxy = self.gradient2(Dx,'y')
Dxz = self.gradient2(Dx,"z")return Dxx, Dyy, Dzz, Dxy, Dxz, Dyz
def eigvalOfhessian2d(self, array):
tmp = np.linalg.eig(array)
lamda =sorted([(abs(tmp[0][0]),tmp[0][0]),(abs(tmp[0][1]),tmp[0][1]),(abs(tmp[0][2]),tmp[0][2])])
Lambda1 = lamda[0][1]
Lambda2 = lamda[1][1]
Lambda3 = lamda[2][1]return Lambda1, Lambda2, Lambda3
def imageEigenvalues(self, I, sigma):
self.hxx, self.hyy,self.hzz, self.hxy, self.hxz, self.hyz= self.Hessian2d(I, sigma)
hxx = self.hxx
hyy = self.hyy
hzz = self.hzz
hxy = self.hxy
hxz = self.hxz
hyz = self.hyz
hxx = hxx.flatten()
hyy = hyy.flatten()
hzz = hzz.flatten()
hxy = hxy.flatten()
hxz = hxz.flatten()
hyz = hyz.flatten()
Lambda1_list =[]
Lambda2_list =[]
Lambda3_list =[]
count =0for i in range(len(hxx)):if hxx[i]+ hyy[i]+ hzz[i]<=0:
Lambda1, Lambda2, Lambda3 =0,0,0else:
array = np.array([[hxx[i],hxy[i],hxz[i]],[hxy[i],hyy[i],hyz[i]],[hxz[i],hyz[i],hzz[i]]])
Lambda1, Lambda2, Lambda3 = self.eigvalOfhessian2d(array)if Lambda1 !=0 and Lambda2!=0 and Lambda3!=0:
count+=1
Lambda1_list.append(Lambda1)
Lambda2_list.append(Lambda2)
Lambda3_list.append(Lambda3)
Lambda1_list = np.array(Lambda1_list)
Lambda2_list = np.array(Lambda2_list)
Lambda3_list = np.array(Lambda3_list)
Lambda1_list[(np.isinf(Lambda1_list))]=0
Lambda2_list[(np.isinf(Lambda2_list))]=0
Lambda3_list[(np.isinf(Lambda3_list))]=0#Lambda1_list[(np.absolute(Lambda1_list)<1e-4)]=0
Lambda1_list = Lambda1_list.reshape(self.size)#Lambda2_list[(np.absolute(Lambda2_list)<1e-4)]=0
Lambda2_list = Lambda2_list.reshape(self.size)#Lambda3_list[(np.absolute(Lambda3_list)<1e-4)]=0
Lambda3_list = Lambda3_list.reshape(self.size)return Lambda1_list,Lambda2_list,Lambda3_list
def vesselness3d(self):for k in range(len(self.sigma)):
lambda1, lambda2, lambda3 = self.imageEigenvalues(self.image, self.sigma[k])
c = self.gaus_image.max()/2
item1 =(1- np.exp(-2*(lambda2 **2)/(lambda3 **2)))
item2 = np.exp(-2*(lambda1 **2)/ np.absolute(lambda2 * lambda3))
item3 =(1- np.exp(-(lambda1 **2+ lambda2 **2+ lambda3 **2)/(2* c **2)))
item1[lambda3==0]=0
item2[lambda3==0]=0
response = item1*item2*item3
response[np.where(np.isnan(response))]=0if k ==0:
vesselness = response
else:
vesselness = np.maximum(vesselness, response)
vesselness =(vesselness /(vesselness.max()))*20000
vesselness[vesselness>200]=200return vesselness
if __name__ =="__main__":
sigma =[0.5,1,1.5,2,2.5]
path ="raw_hepaticvessel_001.nii.gz"
img = sitk.ReadImage(path)
img_data = sitk.GetArrayFromImage(img)
space = img.GetSpacing()
direction = img.GetDirection()
origin = img.GetOrigin()
img_data =200-img_data
v =vesselness3d(img_data,sigma,list(space))
image_data = v.vesselness3d()
img = sitk.GetImageFromArray(image_data)
img.SetOrigin(origin)
img.SetDirection(direction)
img.SetSpacing(space)
sitk.WriteImage(img,"Frangi_hepaticvessel_001.nii.gz")
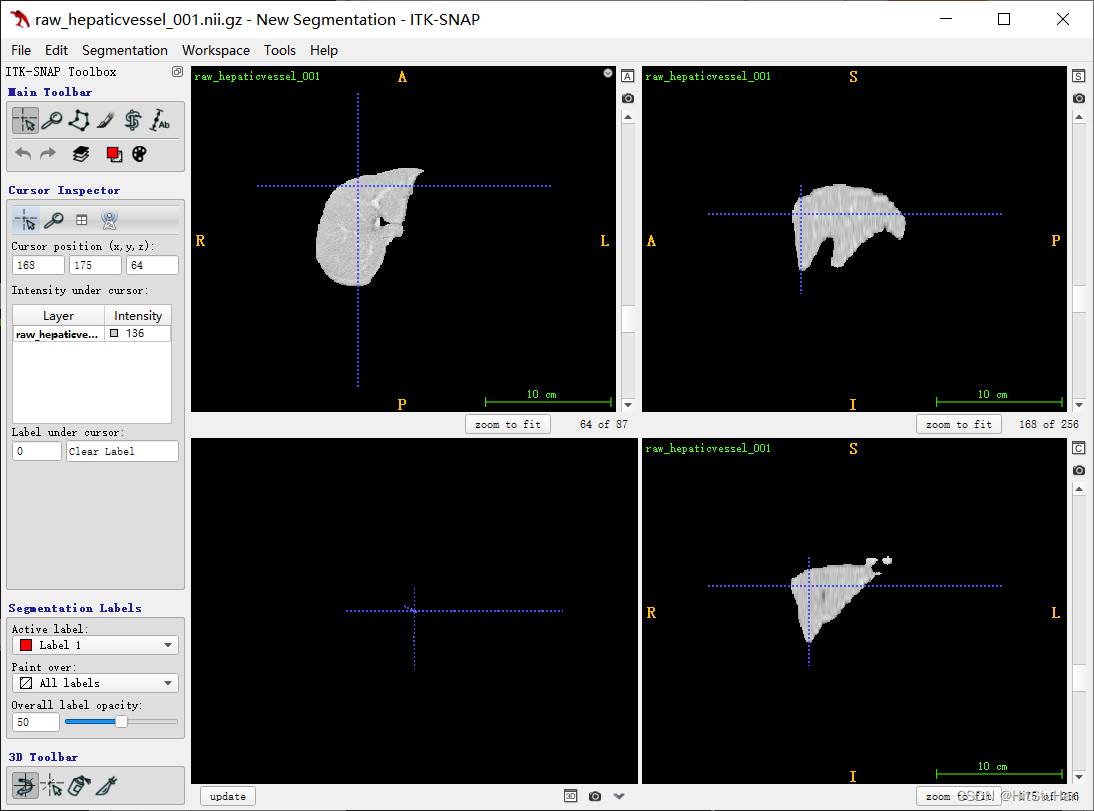
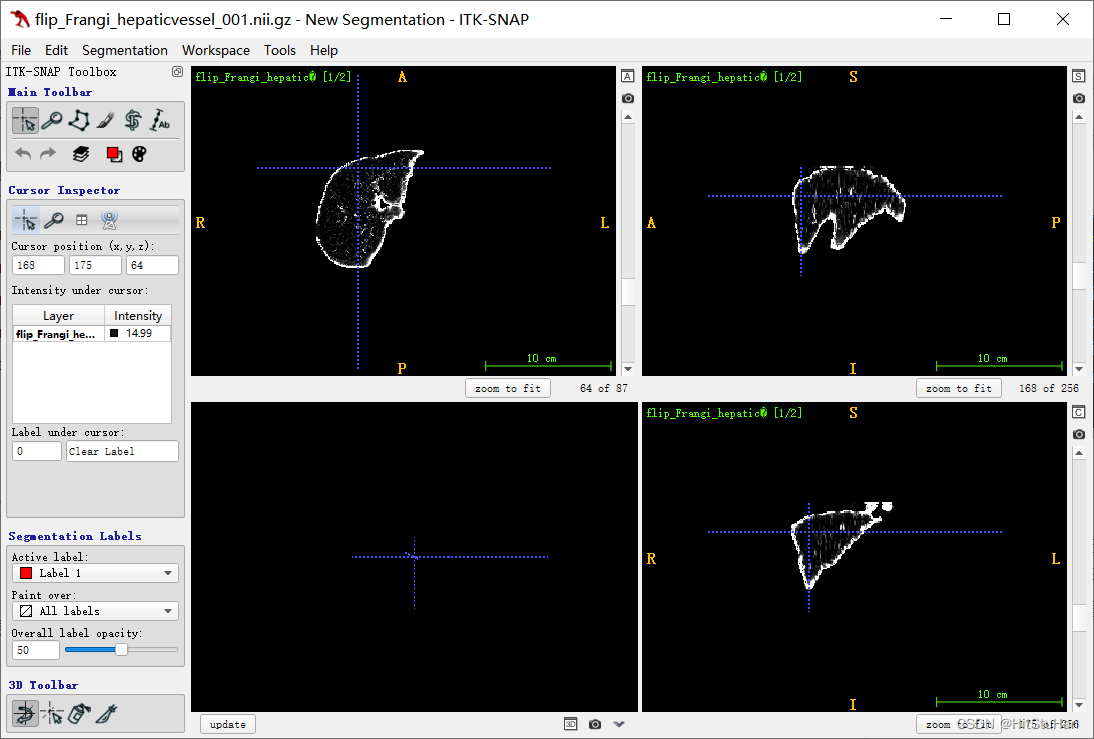
总结
1、Frangi滤波作为经典血管强化、管状强化滤波算法,具有极好的数学证明与实验结果。
2、尽管Frangi滤波效果很好,但仍需要进行调参,如sigma的选取,2D滤波中出现过的tau,已经3D复现中使用的多个超参数。
3、从本文的结论中明显看出,实际上使用三正交平面的滤波效果优于3D Frangi,原因是2中提到的,关于超参的选取问题,而且从2D的复现中,我们能明显看出,代码原作者对Frangi原文做出了极大的改变,使其效果更优。
4、边缘会比血管更容易被增强,所以要处理边缘(在三正交平面我处理了,在3D Frangi没有处理)
5、Frangi滤波的效果告诉我们,机器学习如此发达的今天,特征工程仍必不可少。
版权归原作者 HitStuHan 所有, 如有侵权,请联系我们删除。