Gaussian Naive Bayes (GNB) 是一种基于概率方法和高斯分布的机器学习的分类技术。朴素贝叶斯假设每个参数(也称为特征或预测变量)具有预测输出变量的独立能力。所有参数的预测组合是最终预测,它返回因变量被分类到每个组中的概率,最后的分类被分配给概率较高的分组(类)。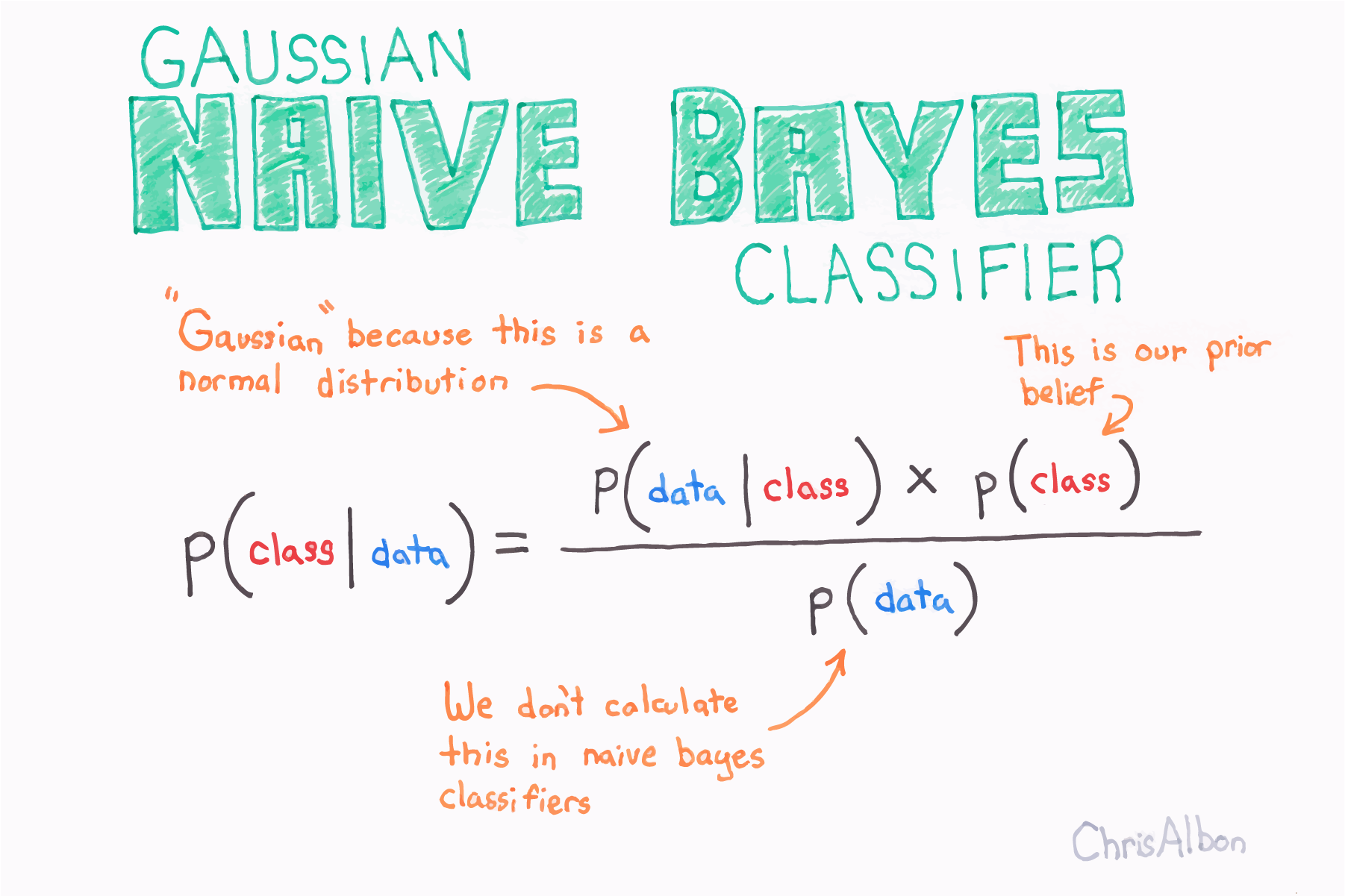
什么是高斯分布?
高斯分布也称为正态分布,是描述自然界中连续随机变量的统计分布的统计模型。正态分布由其钟形曲线定义, 正态分布中两个最重要的特征是均值 (μ) 和标准差 (σ)。平均值是分布的平均值,标准差是分布在平均值周围的“宽度”。
重要的是要知道正态分布的变量 (X) 从 -∞ < X < +∞ 连续分布(连续变量),并且模型曲线下的总面积为 1。
多分类的高斯朴素贝叶斯
导入必要的库:
from random import random
from random import randint
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import statistics
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn.naive_bayes import GaussianNB
from sklearn.metrics import confusion_matrix
from mlxtend.plotting import plot_decision_regions
现在创建一个预测变量呈正态分布的数据集。
#Creating values for FeNO with 3 classes:
FeNO_0 = np.random.normal(20, 19, 200)
FeNO_1 = np.random.normal(40, 20, 200)
FeNO_2 = np.random.normal(60, 20, 200)
#Creating values for FEV1 with 3 classes:
FEV1_0 = np.random.normal(4.65, 1, 200)
FEV1_1 = np.random.normal(3.75, 1.2, 200)
FEV1_2 = np.random.normal(2.85, 1.2, 200)
#Creating values for Broncho Dilation with 3 classes:
BD_0 = np.random.normal(150,49, 200)
BD_1 = np.random.normal(201,50, 200)
BD_2 = np.random.normal(251, 50, 200)
#Creating labels variable with three classes:(2)disease (1)possible disease (0)no disease:
not_asthma = np.zeros((200,), dtype=int)
poss_asthma = np.ones((200,), dtype=int)
asthma = np.full((200,), 2, dtype=int)
#Concatenate classes into one variable:
FeNO = np.concatenate([FeNO_0, FeNO_1, FeNO_2])
FEV1 = np.concatenate([FEV1_0, FEV1_1, FEV1_2])
BD = np.concatenate([BD_0, BD_1, BD_2])
dx = np.concatenate([not_asthma, poss_asthma, asthma])
#Create DataFrame:
df = pd.DataFrame()
#Add variables to DataFrame:
df['FeNO'] = FeNO.tolist()
df['FEV1'] = FEV1.tolist()
df['BD'] = BD.tolist()
df['dx'] = dx.tolist()
#Check database:
df
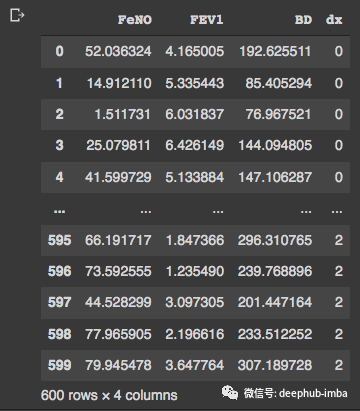
我们的df有 600 行和 4 列。现在我们可以通过可视化检查变量的分布:
fig, axs = plt.subplots(2, 3, figsize=(14, 7))
sns.kdeplot(df['FEV1'], shade=True, color="b", ax=axs[0, 0])
sns.kdeplot(df['FeNO'], shade=True, color="b", ax=axs[0, 1])
sns.kdeplot(df['BD'], shade=True, color="b", ax=axs[0, 2])
sns.distplot( a=df["FEV1"], hist=True, kde=True, rug=False, ax=axs[1, 0])
sns.distplot( a=df["FeNO"], hist=True, kde=True, rug=False, ax=axs[1, 1])
sns.distplot( a=df["BD"], hist=True, kde=True, rug=False, ax=axs[1, 2])
plt.show()
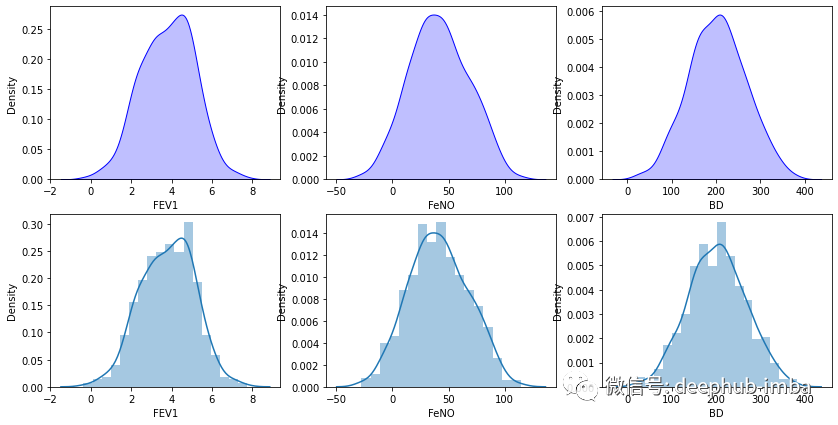
通过人肉的检查,数据似乎接近高斯分布。还可以使用 qq-plots仔细检查:
from statsmodels.graphics.gofplots import qqplot
from matplotlib import pyplot
#q-q plot:
fig, axs = pyplot.subplots(1, 3, figsize=(15, 5))
qqplot(df['FEV1'], line='s', ax=axs[0])
qqplot(df['FeNO'], line='s', ax=axs[1])
qqplot(df['BD'], line='s', ax=axs[2])
pyplot.show()
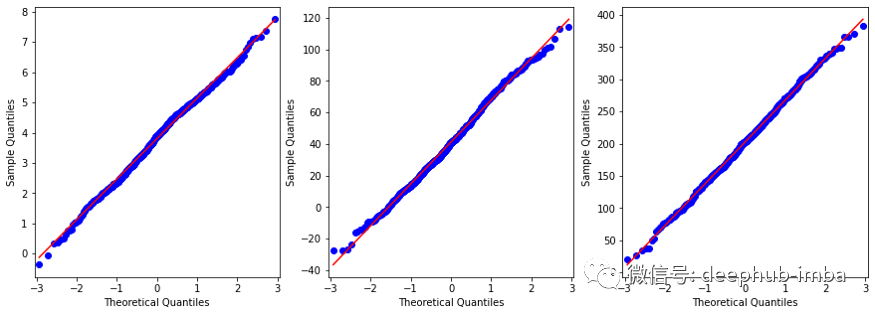
虽然不是完美的正态分布,但已经很接近了。下面查看的数据集和变量之间的相关性:
#Exploring dataset:
sns.pairplot(df, kind="scatter", hue="dx")
plt.show()
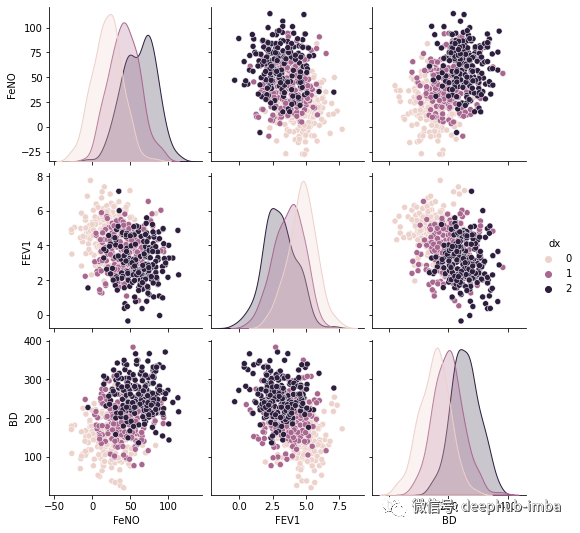
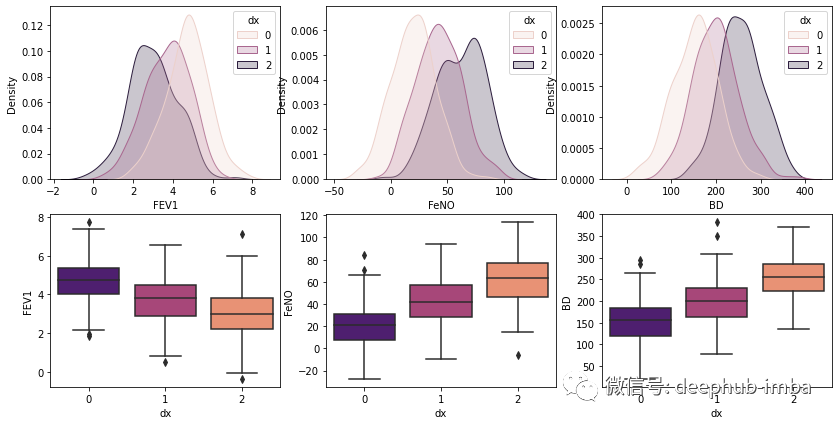
可以使用框线图检查这三组的分布,看看哪些特征可以更好的区分出类别
# plotting both distibutions on the same figure
fig, axs = plt.subplots(2, 3, figsize=(14, 7))
fig = sns.kdeplot(df['FEV1'], hue= df['dx'], shade=True, color="r", ax=axs[0, 0])
fig = sns.kdeplot(df['FeNO'], hue= df['dx'], shade=True, color="r", ax=axs[0, 1])
fig = sns.kdeplot(df['BD'], hue= df['dx'], shade=True, color="r", ax=axs[0, 2])
sns.boxplot(x=df["dx"], y=df["FEV1"], palette = 'magma', ax=axs[1, 0])
sns.boxplot(x=df["dx"], y=df["FeNO"], palette = 'magma',ax=axs[1, 1])
sns.boxplot(x=df["dx"], y=df["BD"], palette = 'magma',ax=axs[1, 2])
plt.show()
手写朴素贝叶斯分类
手写代码并不是让我们重复的制造轮子,而是通过自己编写代码对算法更好的理解。在进行贝叶斯分类之前,先要了解正态分布。
正态分布的数学公式定义了一个观测值出现在某个群体中的概率:
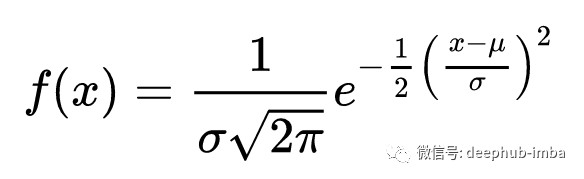
我们可以创建一个函数来计算这个概率:
def normal_dist(x , mean , sd):
prob_density = (1/sd*np.sqrt(2*np.pi)) * np.exp(-0.5*((x-mean)/sd)**2)
return prob_density
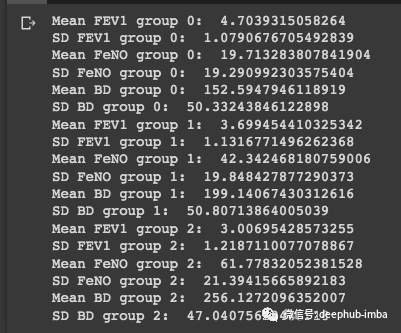
知道正态分布公式,就可以计算该样本在三个分组(分类)概率。首先,需要计算所有预测特征和组的均值和标准差:
#Group 0:
group_0 = df[df['dx'] == 0]print('Mean FEV1 group 0: ', statistics.mean(group_0['FEV1']))
print('SD FEV1 group 0: ', statistics.stdev(group_0['FEV1']))
print('Mean FeNO group 0: ', statistics.mean(group_0['FeNO']))
print('SD FeNO group 0: ', statistics.stdev(group_0['FeNO']))
print('Mean BD group 0: ', statistics.mean(group_0['BD']))
print('SD BD group 0: ', statistics.stdev(group_0['BD']))
#Group 1:
group_1 = df[df['dx'] == 1]
print('Mean FEV1 group 1: ', statistics.mean(group_1['FEV1']))
print('SD FEV1 group 1: ', statistics.stdev(group_1['FEV1']))
print('Mean FeNO group 1: ', statistics.mean(group_1['FeNO']))
print('SD FeNO group 1: ', statistics.stdev(group_1['FeNO']))
print('Mean BD group 1: ', statistics.mean(group_1['BD']))
print('SD BD group 1: ', statistics.stdev(group_1['BD']))
#Group 2:
group_2 = df[df['dx'] == 2]
print('Mean FEV1 group 2: ', statistics.mean(group_2['FEV1']))
print('SD FEV1 group 2: ', statistics.stdev(group_2['FEV1']))
print('Mean FeNO group 2: ', statistics.mean(group_2['FeNO']))
print('SD FeNO group 2: ', statistics.stdev(group_2['FeNO']))
print('Mean BD group 2: ', statistics.mean(group_2['BD']))
print('SD BD group 2: ', statistics.stdev(group_2['BD']))
现在,使用一个随机的样本进行测试:FEV1 = 2.75FeNO = 27BD = 125
#Probability for:
#FEV1 = 2.75
#FeNO = 27
#BD = 125
#We have the same number of observations, so the general probability is: 0.33
Prob_geral = round(0.333, 3)
#Prob FEV1:
Prob_FEV1_0 = round(normal_dist(2.75, 4.70, 1.08), 10)
print('Prob FEV1 0: ', Prob_FEV1_0)
Prob_FEV1_1 = round(normal_dist(2.75, 3.70, 1.13), 10)
print('Prob FEV1 1: ', Prob_FEV1_1)
Prob_FEV1_2 = round(normal_dist(2.75, 3.01, 1.22), 10)
print('Prob FEV1 2: ', Prob_FEV1_2)
#Prob FeNO:
Prob_FeNO_0 = round(normal_dist(27, 19.71, 19.29), 10)
print('Prob FeNO 0: ', Prob_FeNO_0)
Prob_FeNO_1 = round(normal_dist(27, 42.34, 19.85), 10)
print('Prob FeNO 1: ', Prob_FeNO_1)
Prob_FeNO_2 = round(normal_dist(27, 61.78, 21.39), 10)
print('Prob FeNO 2: ', Prob_FeNO_2)
#Prob BD:
Prob_BD_0 = round(normal_dist(125, 152.59, 50.33), 10)
print('Prob BD 0: ', Prob_BD_0)
Prob_BD_1 = round(normal_dist(125, 199.14, 50.81), 10)
print('Prob BD 1: ', Prob_BD_1)
Prob_BD_2 = round(normal_dist(125, 256.13, 47.04), 10)
print('Prob BD 2: ', Prob_BD_2)
#Compute probability:
Prob_group_0 = Prob_geral*Prob_FEV1_0*Prob_FeNO_0*Prob_BD_0
print('Prob group 0: ', Prob_group_0)
Prob_group_1 = Prob_geral*Prob_FEV1_1*Prob_FeNO_1*Prob_BD_1
print('Prob group 1: ', Prob_group_1)
Prob_group_2 = Prob_geral*Prob_FEV1_2*Prob_FeNO_2*Prob_BD_2
print('Prob group 2: ', Prob_group_2)
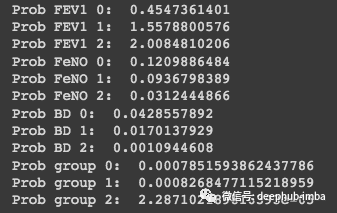
可以看到,这个样本具有属于第 2 组的概率最高。这就是朴素贝叶斯手动计算的的流程,但是这种成熟的算法可以使用来自 Scikit-Learn 的更高效的实现。
Scikit-Learn的分类器样例
Scikit-Learn的GaussianNB为我们提供了更加高效的方法,下面我们使用GaussianNB进行完整的分类实例。首先创建 X 和 y 变量,并执行训练和测试拆分:
#Creating X and y:
X = df.drop('dx', axis=1)
y = df['dx']
#Data split into train and test:
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.30)
在输入之前还需要使用 standardscaler 对数据进行标准化:
sc = StandardScaler()
X_train = sc.fit_transform(X_train)
X_test = sc.transform(X_test)
现在构建和评估模型:
#Build the model:
classifier = GaussianNB()
classifier.fit(X_train, y_train)
#Evaluate the model:
print("training set score: %f" % classifier.score(X_train, y_train))
print("test set score: %f" % classifier.score(X_test, y_test))
下面使用混淆矩阵来可视化结果:
# Predicting the Test set results
y_pred = classifier.predict(X_test)
#Confusion Matrix:
cm = confusion_matrix(y_test, y_pred)
print(cm)
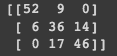
通过混淆矩阵可以看到,的模型最适合预测类别 0,但类别 1 和 2 的错误率很高。为了查看这个问题,我们使用变量构建决策边界图:
df.to_csv('data.csv', index = False)
data = pd.read_csv('data.csv')
def gaussian_nb_a(data):
x = data[['BD','FeNO',]].values
y = data['dx'].astype(int).values
Gauss_nb = GaussianNB()
Gauss_nb.fit(x,y)
print(Gauss_nb.score(x,y))
#Plot decision region:
plot_decision_regions(x,y, clf=Gauss_nb, legend=1)
#Adding axes annotations:
plt.xlabel('X_train')
plt.ylabel('y_train')
plt.title('Gaussian Naive Bayes')
plt.show()
def gaussian_nb_b(data):
x = data[['BD','FEV1',]].values
y = data['dx'].astype(int).values
Gauss_nb = GaussianNB()
Gauss_nb.fit(x,y)
print(Gauss_nb.score(x,y))
#Plot decision region:
plot_decision_regions(x,y, clf=Gauss_nb, legend=1)
#Adding axes annotations:
plt.xlabel('X_train')
plt.ylabel('y_train')
plt.title('Gaussian Naive Bayes')
plt.show()
def gaussian_nb_c(data):
x = data[['FEV1','FeNO',]].values
y = data['dx'].astype(int).values
Gauss_nb = GaussianNB()
Gauss_nb.fit(x,y)
print(Gauss_nb.score(x,y))
#Plot decision region:
plot_decision_regions(x,y, clf=Gauss_nb, legend=1)
#Adding axes annotations:
plt.xlabel('X_train')
plt.ylabel('y_train')
plt.title('Gaussian Naive Bayes')
plt.show()
gaussian_nb_a(data)
gaussian_nb_b(data)
gaussian_nb_c(data)
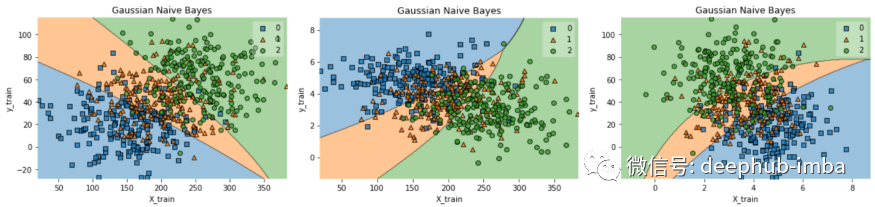
通过决策边界我们可以观察到分类错误的原因,从图中我们看到,很多点都是落在决策边界之外的,如果是实际数据我们需要分析具体原因,但是因为是测试数据所以我们也不需要更多的分析。