基于LIDC-IDRI肺结节肺癌数据集的人工智能深度学习分类良性和恶性肺癌(Python 全代码)全流程解析(二)
第一部分内容的传送门
第三部分传送门
1 环境配置和数据集预处理
1.1 环境配置
环境配置建议使用anaconda进行配置。核心的配置是keras和tensorflow的版本要匹配。
环境配置如下:
tensorboard 1.13.1
tensorflow 1.13.1
Keras 2.2.4
numpy 1.21.5
opencv-python 4.6.0.66
python 3.7
1.1 数据集预处理
数据集的预处理分为两个关键步骤。首先是图片处理,我们使用cv2库将图片转换为矩阵格式。这些矩阵随后被堆叠,并放入一个列表中,以便于深度学习模型的读取和处理。
其次是标签处理。我们从保存肺癌恶性程度信息的label.csv文件中逐行读取数据。通过切片和提取,我们获取了肺癌的恶性程度评级,这些评级在1到5之间。我们将大于3的评级归类为恶性,小于3的评级归类为良性。为了让模型更好地理解这些标签,我们用1表示良性,0表示恶性,最后将标签数据转换为one-hot编码格式。
输出与处理的函数如下:
import matplotlib.pyplot as plt
import numpy as np
import keras
import cv2
import os
from keras.preprocessing.image import img_to_array
from keras.utils import to_categorical, plot_model
defload_data(label_path,data_path):
data_x =[]
labels =[]
f =open(label_path)
label = f.readlines()for Pathimg in os.listdir(os.path.join(data_path,'x')):
Path = os.path.join(os.path.join(data_path,'x'),Pathimg)#print(Path)
image = cv2.imread(Path)
image = img_to_array(image)
data_x.append(image)#处理label
index_num =int(Pathimg.split('.')[0])# print(labels)用索隐处理,
a = label[index_num]
label_ =int(a[-3:-2])
label_1 =1if label_ >3else0
labels.append(label_1)print(labels)# print(data)#guiyihua
data_x = np.array(data_x,dtype='float')/255.0
labels = np.array(labels)#转化标签为张量
labels = to_categorical(labels)#载入data——y
data_y =[]for Pathimg in os.listdir(os.path.join(data_path,'y')):
Path = os.path.join(os.path.join(data_path,'y'),Pathimg)#print(Path)
image = cv2.imread(Path)
image = img_to_array(image)
data_y.append(image)#guiyihua
data_y = np.array(data_y,dtype='float')/255.0#处理Z
data_z =[]for Pathimg in os.listdir(os.path.join(data_path,'z')):
Path = os.path.join(os.path.join(data_path,'z'),Pathimg)#print(Path)
image = cv2.imread(Path)
image = img_to_array(image)
data_z.append(image)#guiyihua
data_z = np.array(data_y,dtype='float')/255.0return labels,data_x,data_y,data_z
我们定义好数据预处理的函数后读取数据。图片的数据的格式如下:
其中有875个图片,每个图片的大小为50*50和3个通道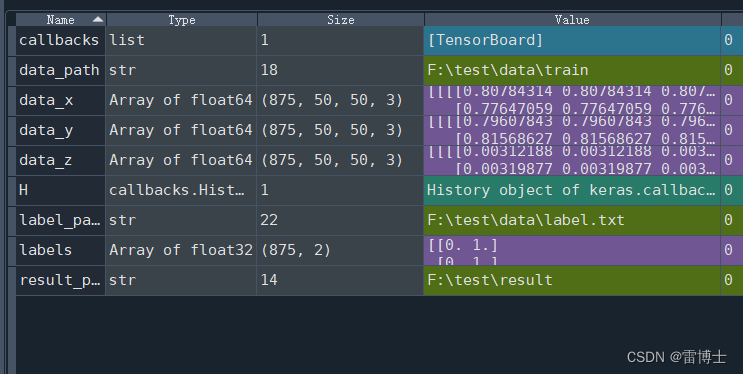
label的格式如下: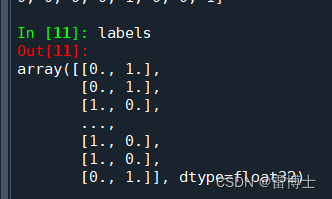
2 深度学习模型训练和评估
2.1 深度学习模型训练
我们已经在另一个文件中创建了一个深度学习模型,并且定义了一个函数来导入这个模型。在这里,我们将实例化这个函数。该函数会返回模型的结构以及模型训练的记录信息。
这个模型的输入是来自三个不同角度的图像和对应的标签。模型将被训练 5 个周期,每个周期训练 50 个图像。我们将使用 TensorBoard 查看模型的训练记录。
#######搭建模型
model,callbacks = create_model()#模型训练
H = model.fit([data_x,data_y,data_z],[labels],
epochs=5,batch_size=50, callbacks=callbacks)
训练结果如下:
我们可以看到训练的准确率还可以为0.7451左右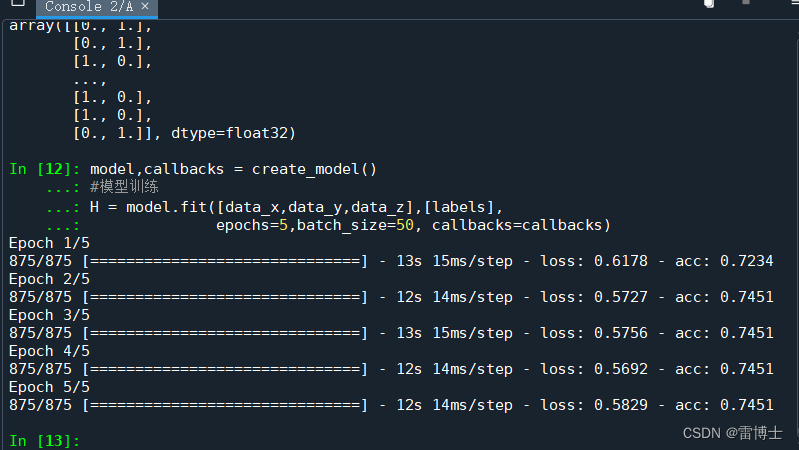
训练完成后我们将模型储存起来方便对模型进行评估。
#######save model to diskprint('info:saving model.....')
model.save(os.path.join(result_path,'model_44.h5'))
2.1 深度学习模型评估
模型的评估我们首先使用了,分类模型训练的损失函数和准确率来评估模型的训练过程。之后我们还会介绍模型的评估的其他指标。包括混淆矩阵,ROC曲线,AUC值,等
#准确率损失曲线绘制
plt.style.use('ggplot')
plt.figure()
N =5
fig = plt.plot(np.arange(0, N), H.history["loss"], label="train_loss")#plt.plot(np.arange(0, N), H.history["val_loss"], label="val_loss")
plt.plot(np.arange(0, N), H.history["acc"], label="train_acc")#plt.plot(np.arange(0, N), H.history["val_acc"], label="val_acc")
plt.title("Training Loss and Accuracy on traffic-sign classifier")
plt.xlabel("Epoch #")
plt.ylabel("Loss/Accuracy")
plt.legend(loc="lower left")#plt.show()
plt.savefig('a.jpg',dpi=800)
训练结果如下: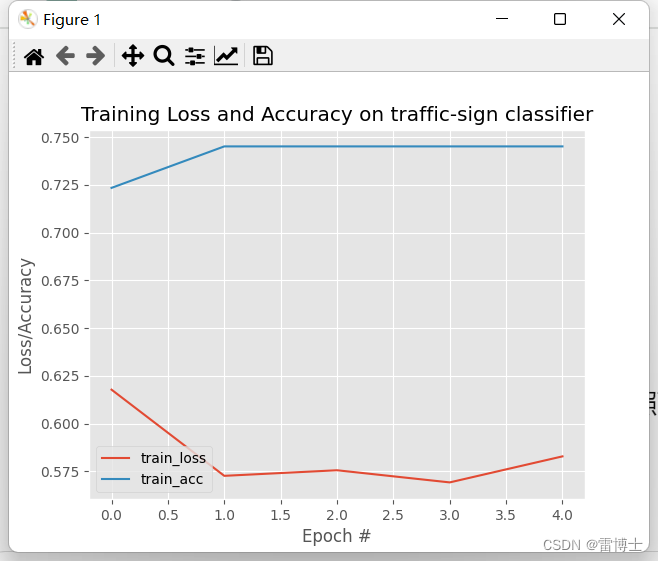
笑话一则开心一下喽
今天去买水果,我问一个摊主:别人家的瓜都写着不甜包退,你怎么不敢写呢,瓜不好吧。摊主:不买滚,我他么卖的是苦瓜……
深度学习,医学图像处理,机器学习精通,需要帮助的联系我(有偿哦)
完整代码如下:
import matplotlib.pyplot as plt
import numpy as np
import keras
import cv2
import os
os.chdir('F:\工作\博客\sort_lung')from models import create_model
from load_datas import load_data
label_path =r'F:\test\data\label.txt'
data_path =r'F:\test\data\train'
result_path =r'F:\test\result'#读取预处理数据
labels,data_x,data_y,data_z = load_data(label_path,data_path)#######搭建模型
model,callbacks = create_model()#模型训练
H = model.fit([data_x,data_y,data_z],[labels],
epochs=5,batch_size=50, callbacks=callbacks)#save model to diskprint('info:saving model.....')
model.save(os.path.join(result_path,'model_44.h5'))#准确率损失曲线绘制
plt.style.use('ggplot')
plt.figure()
N =5
fig = plt.plot(np.arange(0, N), H.history["loss"], label="train_loss")#plt.plot(np.arange(0, N), H.history["val_loss"], label="val_loss")
plt.plot(np.arange(0, N), H.history["acc"], label="train_acc")#plt.plot(np.arange(0, N), H.history["val_acc"], label="val_acc")
plt.title("Training Loss and Accuracy on traffic-sign classifier")
plt.xlabel("Epoch #")
plt.ylabel("Loss/Accuracy")
plt.legend(loc="lower left")#plt.show()
plt.savefig('a.jpg',dpi=800)
模型文件如下
深度学习模型讲解—待续
# -*- coding: utf-8 -*-"""
Created on Sun Apr 14 21:08:01 2024
@author: dell
"""from keras.preprocessing.image import ImageDataGenerator
from keras.optimizers import Adam
from sklearn.model_selection import train_test_split
from keras.preprocessing.image import img_to_array
from keras.utils import to_categorical, plot_model
from keras.models import Model
#from imutils import pathsimport matplotlib.pyplot as plt
import numpy as np
import keras
import cv2
import os
#定义模型from keras.models import Sequential
from keras.layers import Conv2D, MaxPooling2D
from keras.layers import Activation, Dropout, Flatten, Dense, Input,Concatenate
defcreate_model():
model = Sequential()#############定义多输入
input1 = Input(shape=(50,50,3),name ='input1')
input2 = Input(shape=(50,50,3),name ='input2')
input3 = Input(shape=(50,50,3),name ='input3')#############定义多输入
x1 = Conv2D(32,(3,3),padding='same')(input1)#input is height,width,deep
x1 = Activation('relu')(x1)
x1 = Conv2D(32,(3,3),padding='same')(x1)
x1 = Activation('relu')(x1)
x1 = MaxPooling2D(pool_size=(2,2),strides =(2,2))(x1)
x1 = Conv2D(48,(3,3),padding='same')(x1)
x1 = Activation('relu')(x1)
x1 = Conv2D(48,(3,3),padding='same')(x1)
x1 = Activation('relu')(x1)
x1 = MaxPooling2D(pool_size=(2,2),strides =(2,2))(x1)
x1 = Conv2D(64,(3,3),padding='same')(x1)
x1 = Activation('relu')(x1)
x1 = Conv2D(64,(3,3),padding='same')(x1)
x1 = Activation('relu')(x1)
x1 = MaxPooling2D(pool_size=(2,2),strides =(2,2))(x1)# the model so far outputs 3D feature maps (height, width, features)#base_model.add(Flatten()) # this converts our 3D feature maps to 1D feature vectors
x1 = Flatten()(x1)
x1 = Dense(256)(x1)
x1 = Activation('relu')(x1)
x1 = Dropout(0.5)(x1)
category_predict1 = Dense(100, activation='softmax', name='category_predict1')(x1)# Three loss functions#定义三个全连接层
x2 = Conv2D(32,(3,3),padding='same')(input2)#input is height,width,deep
x2 = Activation('relu')(x2)
x2 = Conv2D(32,(3,3),padding='same')(x2)
x2 = Activation('relu')(x2)
x2 = MaxPooling2D(pool_size=(2,2),strides =(2,2))(x2)
x2 = Conv2D(48,(3,3),padding='same')(x2)
x2 = Activation('relu')(x2)
x2 = Conv2D(48,(3,3),padding='same')(x2)
x2 = Activation('relu')(x2)
x2 = MaxPooling2D(pool_size=(2,2),strides =(2,2))(x2)
x2 = Conv2D(64,(3,3),padding='same')(x2)
x2 = Activation('relu')(x2)
x2 = Conv2D(64,(3,3),padding='same')(x2)
x2 = Activation('relu')(x2)
x2 = MaxPooling2D(pool_size=(2,2),strides =(2,2))(x2)# the model so far outputs 3D feature maps (height, width, features)#base_model.add(Flatten()) # this converts our 3D feature maps to 1D feature vectors
x2 = Flatten()(x2)
x2 = Dense(256)(x2)
x2 = Activation('relu')(x2)
x2 = Dropout(0.5)(x2)
category_predict2 = Dense(100, activation='relu', name='category_predict2')(x2)
x3 = Conv2D(32,(3,3),padding='same')(input3)#input is height,width,deep
x3 = Activation('relu')(x3)
x3 = Conv2D(32,(3,3),padding='same')(x3)
x3 = Activation('relu')(x3)
x3 = MaxPooling2D(pool_size=(2,2),strides =(2,2))(x3)
x3 = Conv2D(48,(3,3),padding='same')(x3)
x3 = Activation('relu')(x3)
x3 = Conv2D(48,(3,3),padding='same')(x3)
x3 = Activation('relu')(x3)
x3 = MaxPooling2D(pool_size=(2,2),strides =(2,2))(x3)
x3 = Conv2D(64,(3,3),padding='same')(x3)
x3 = Activation('relu')(x3)
x3 = Conv2D(64,(3,3),padding='same')(x3)
x3 = Activation('relu')(x3)
x3 = MaxPooling2D(pool_size=(2,2),strides =(2,2))(x3)# the model so far outputs 3D feature maps (height, width, features)#base_model.add(Flatten()) # this converts our 3D feature maps to 1D feature vectors
x3 = Flatten()(x3)
x3 = Dense(256)(x3)
x3 = Activation('relu')(x3)
x3 = Dropout(0.5)(x3)
category_predict3 = Dense(100, activation='relu', name='category_predict3')(x3)#融合全连接层
merge = Concatenate()([category_predict1,category_predict2,category_predict3])#定义输出
output = Dense(2,activation='sigmoid', name='output')(merge)
model = Model(inputs=[input1, input2, input3], outputs=[output])
callbacks =[keras.callbacks.TensorBoard(
log_dir='my_log_dir',)]
model.compile(optimizer=Adam(lr=0.001,decay=0.01),
loss='binary_crossentropy',
metrics=['accuracy'],)return model,callbacks
版权归原作者 雷博士 所有, 如有侵权,请联系我们删除。